This is used in generating diversity in models. Apart from mutations, other types of drug resistance mechanisms, e. This can be helpful on NFS-mounted systems when running with multiple processors to avoid file conflicts. Anchor-and-Grow based docking program. All three versions are identical except in the way that they take commands from the user. 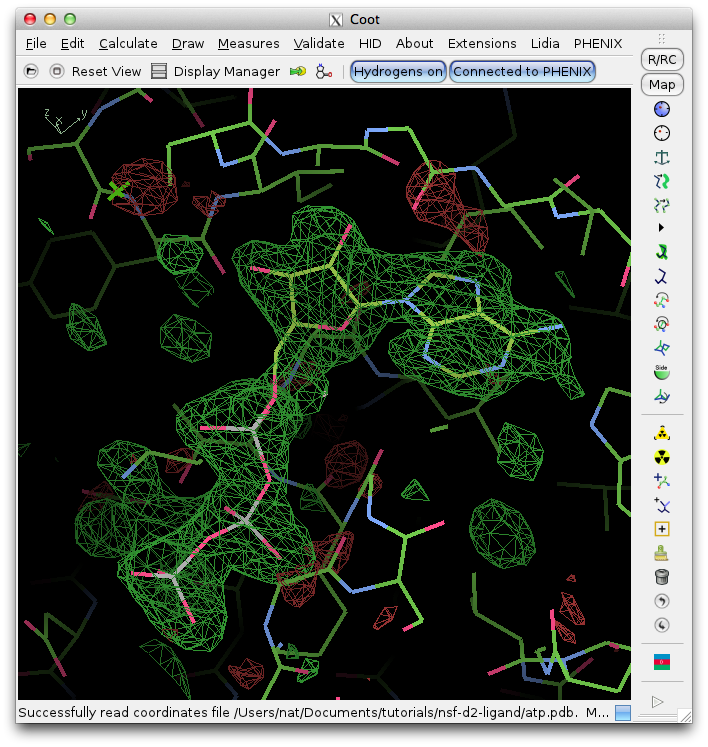
| Uploader: | Fenrijind |
| Date Added: | 20 August 2007 |
| File Size: | 36.62 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 33761 |
| Price: | Free* [*Free Regsitration Required] |
You can upload a protein and a docking is performed, either against an in-house database containing more than a million active compounds, or against a user-defined library. However, no method was consistently superior to other approaches across different data sets. Open for general research.
Maximum-Entropy based docking web server for efficient prediction of ligand binding sites. The Protein Model Portal—a comprehensive resource for protein structure and model information.
While these techniques relying on 1D- and 2D-descriptors are suitable for screening millions of compounds, more complex scoring functions using 3D- and 4D-descriptors, statistical learning and available binding affinities for already known binders can provide more accurate estimations, but involve significantly higher runtimes i. To collect information on the known protein—ligand interactions for a receptor or small molecule of interest, Table 2 lists the main relevant databases, most of which are publicly accessible.
Flexible ligand docking to multiple receptor conformations: A high-throughput screening study applied to the thymidine kinase receptor is also presented in which LigandFit, when combined with LigScore, an internally developed scoring function, yields very good hit rates for a ligand pool seeded with known actives.
Multiple Structural Alignments are built on the fly within LigBase from a series of pairwise alignments. The scoring method to quantify the structural similarity mainly depends on the used descriptor types and individual choices on how to weigh the relevance of different molecular features. The easiest way to do this is to select highlight a ligand on the Results page in the GUI and click on one of the display options provided.

Pharmacophore identification by hypermolecular alignment of ligands in 3D. Web-driven interface for performing structure-based virtual screening with AutoDock Vina. Protein softwarre overlapping with the ligand density will prevent a successful placement, so some editing of the input structure is generally recommended.
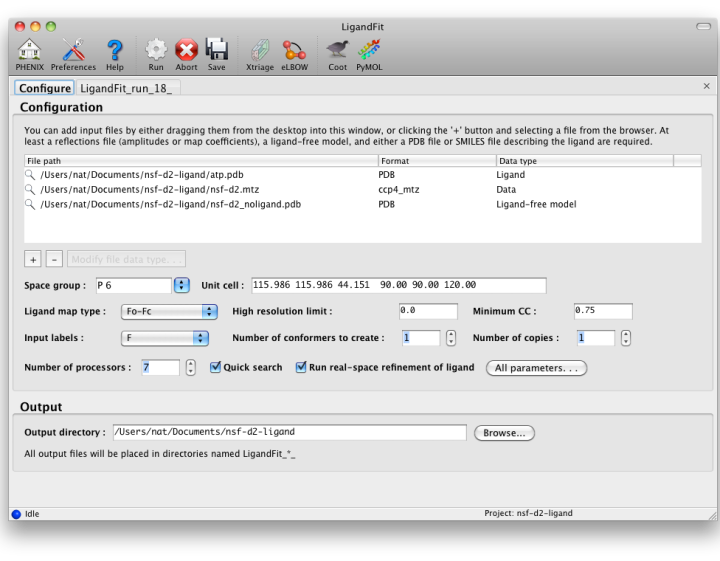
A value between 0. A unified, probabilistic framework for structure- and ligand-based virtual screening. Although this task still remains a challenge, dedicated approaches are available, e. Output files from LigandFit When you run LigandFit the output files will be ligandvit a subdirectory with your run number: A PDB file containing the initial ligand-free model.
A novel high-frequency sub-pathway mining approach to identify robust prognostic gene signatures. Only applies to density modified maps.
Furthermore, interaction-based classifier, trained on a target-specific knowledge base can be used in a post-docking filter step. Progam for similarity-based docking, based on a combination of the ligand based GMA molecular alignment tool and the docking tool GlamDock. Mean AUC values between 0.
Using the LigandFit GUI
In summary, softare management and virtualization tools provide new means to obtain reproducible and portable screening pipelines, which can be adjusted and extended with minimal effort. The web site can be downloaded and installed independently from GitHub. Ligand binding site prediction from protein sequence and structure.
You can email us confidentially at help phenix-online. Docking program based on an idealized active site ligand a protomolused as a target to generate putative poses of molecules or molecular fragments, which are scored using the Hammerhead scoring function.
Frequently asked questions about ligands
Exists as a standalone program. Prediction tool for protein binding sites. Automated method for the prediction of ligand binding sites.
List of molecular graphics systems.
Click2Drug
If you encounter especially problematic ligands, we are interested in examining these, as they occasionally expose bugs. Nucleos identifies binding sites for nucleotide modules namely the nucleobase, the sofyware and the phosphate and then combines them in order to build the complete binding sites for different types of oigandfit e. A methodology tehat predicts pocket druggability, efficient on both; estimated pockets guided by the ligand proximity extracted by proximity to a ligand from a holo protein structure using several thresholds and estimated pockets not guided by the ligand proximity based on amino atoms that form the surface of potential binding cavities.

No comments:
Post a Comment